This post is about the extraordinary lengths one must go to, in order to get a journal or institution to take action on a published paper that contains problematic data. This has been a long time in the making, and is important (to me at least) because it’s both a last ditch attempt to get something done, and the first time I’ve used this website as a forum for such material. The post is in 3 parts: (i) A detailed description of the problems with the paper itself. (ii) A narrative of my attempts to get the scientific record corrected. (iii) Some concluding thoughts on the sorry state of affairs in academic publishing today.
The paper in question is this one… PLoS Biology (2013) 11, e1001603. Effects of Resveratrol and SIRT1 on PGC-1α Activity and Mitochondrial Biogenesis: A Reevaluation. [PMID 23874150]
Part 1 – The Problem
I first came across this paper because it featured prominently in a New York Times article entitled “Exercise in a Pill“. I work in the area of SIRT1 and mitochondrial biology, so I thought surely a paper on these topics featured in NYT must be worth a read. However, upon getting into the paper I rapidly discovered some potential problems with the data – it appeared some of the western blots may have been re-used between different figures, and some other data didn’t follow “best practices” that would be necessary given the bold claims being made. The following images are included to illustrate the some of these anomalies.
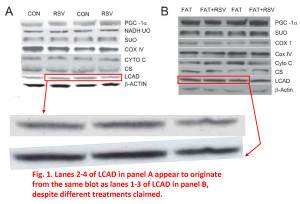
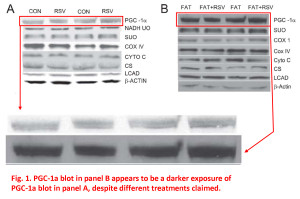
In these first 2 examples from Figure 1, it appears as if some of the blot images in panel A and panel B are simply different exposures of the same image, but they’re used to represent different samples, different experimental conditions. The patterns of the bands, their juxtaposition, their shape, various imperfections and spots, all fall into the category of “more similar than would be expected purely by chance”.
The same appears here in Figure 6 (above), in which 2 blots one on top of the other appear to be different exposures of the same image. This one is tricky to pin down, because the 2 proteins of interest (COX IV and cytochrome c) have very similar apparent molecular weights (~15 kDa) on SDS-PAGE, so it’s possible that if the same membrane was used to blot for these 2 proteins, with a strip/re-probe in between, then the same imperfections would come through in the final blot. The problem is, even if this perfectly good explanation is in-fact the case, it begs the question why would you strip and re-probe for 2 proteins that run at the same weight on a blot? There are some situations in which this is permitted or encouraged (e.g., if you want to measure the phosphorylation status of a protein, you probe with the phospho-X antibody, then strip the blot at re-probe with the total-X antibody, and normalize the former to the latter – see below). However, when dealing with two separate proteins at almost identical mass, using the same blot twice definitely falls outside of best practices.
In Figure 4, the phospho-versus-total scenario comes up. As described above, the typical way these experiments work is to probe with the phospho-X antibody, then strip the blot and re-probe with the total-X antibody, then normalize phospho-X to total-X. When this is done, we can be sure that the phospho-protein signal actually changed, because the normalization (the total protein) is right there on the same gel. However, in this case the phospho blots have been spliced together, as indicated by the vertical lines in the panels, but the total protein blots have not been spliced. In-fact, in the top panel, that blot image is 4 separate bands pasted together from 4 separate blots. Sometimes it happens – you run the samples in the wrong order and you have to rearrange the samples to get them in the “right” order for publishing (IMHO this is lazy, just run the blot again the way you want it to be). If this was the case, wouldn’t the samples be mixed up on all the gels for a particular experiment (assuming they’re all run at the same time)? What we’re asked to believe here, is that the authors ran the phospho-AMPK blot with the samples in one order, and the total-AMPK blot with the samples in another order (the correct one, it seems) and then they only rearranged one set of bands so they all matched up. Needless to say, the potential for “convenient adjustments” to the data during this rearrangement, is not a factor in pure un-spliced blots.
The same then happens in the lower 2 panels for phospho-ACC versus total-ACC (i.e. one is spliced, the other not). But, this one has another problem too… generally speaking phospho protein is a sub-set of total protein. So, you might find that a total-protein antibody will recognize multiple bands (maybe different isoforms of the protein), but then the phospho-specific antibody should be more… well… specific. The odd thing here, is the p-ACC blot has 2 bands, but the total ACC blot (which should include all the phosphorylated and other ACC forms) only has a single band. This falls in the “just plain weird” category. There’s also the glaring problem that the shapes/positioning of all the bands between phospho-X and total-X just don’t match up. Clearly these blots originated from different membranes, and while this is not strictly forbidden, it certainly doesn’t count as best practices either.
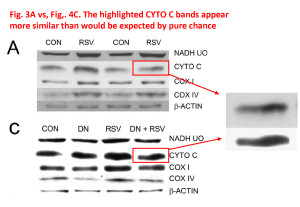
Finally, there’s this anomaly between Figures 3A and 4C. It’s hard to be sure, but to me those bands just look too similar, with one possibly being a grossly overexposed version of the other. What’s interesting also, is the lower one is pure black on a pure white background, which makes it impossible to “anchor” the band to its surroundings. This happens a lot in blots presented in papers – people crank up the contrast and adjust the brightness so their blots appear black/white instead of dark-gray/light-gray. It might conform to what some people think a blot should look like, but it also introduces the potential for hiding splices which is not there in a grayscale image. It’s impossible to tell if a band is spliced because when you splice together pure white and pure white there’s no seam.
This does impact the interpretation, because blot densitometry relies on the bands being within the linear part of the dynamic range, wherein black = 100% and white = 0%. As such, any band on a western lot in which the center is completely saturated black is unsuitable for densitometry (it will yield a tableau profile, a clipped peak, instead of a nice Gaussian peak). Thus, it’s not really possible to believe any quantitative data originating from such blots.
In addition to the above, the paper contains 85 panels of western blot data, and every single one is cropped (“letterboxed”) and presented without any molecular weight markers. A lot of them are over-saturated and unsuitable for densitometry, or contain pure black/white bands which make it impossible to tell if they’ve been spliced. There are also instances in which the same antibody in the same cells recognized 1 band in one panel and 3 bands in a different data panel (why so much variability in what the antibody recognizes?) Altogether, there are a host of problems with the data in this paper, and while they might all have a perfectly good explanation, at the very least some of these things fall into the column of “makes me question the conclusions”. So, what did I do about it?
Part 2 – The Solution (I thought!)
Thankfully, the InterNet has afforded a number of tools enabling readers to comment on the published literature and (one hopes) correct the scientific record. In addition to comment systems at individual journals, sites such as PubPeer and PubMed Commons are pushing the boundaries of Post Publication Peer Review even further, aiming for centralized discussion forums. This is a good thing. We can all look forward to the day when the integrity of a paper is not judged on the basis of the impact factor of the journal it’s published in, but on the quality and reproducibility of the data inside!
So, having read this paper, I trotted on over to the PLoS Biology website and left a comment, outlining the problems above. Here is a saved (PDF) copy of the comment (July 18th). I even used my real name! I also Tweeted Gretchen Reynolds, the NYT reporter who wrote the piece about Exercise in a Pill. She didn’t respond.
You might ask why I can’t just provide a link to the comment itself. Well herein lies the problem – PLoS Biology decided that it violated their terms of service and deleted it. The email conversation went like this:
PLoS: We’d like to remove your comment because we believe it violates our terms.
Me: I don’t think it does. Read the disclaimer I wrote at the end of my comment.
Comment removed
Me: What just happened? Can you explain what you plan to do about this? Why did you act unilaterally and not allow me to explain why my comment is valid?
Me: Hello, anybody home? Oh, I see, you’re just ignoring me now.
Silence. Nothing beyond the boilerplate that they take comments seriously and will “discuss this with the authors”. I even took pains to remind them of the Streisand Effect, whereupon removing the comment might actually have the opposite of the desired effect. Just for fun, let’a take a look at the PLoS commenting terms and conditions. #7 caught my eye…
Questions about experimental data are appropriate, but need to be phrased in a way that does not imply any misconduct on the part of the authors.
Now go read the comment again, particularly my lengthy disclaimer at the end, in which I specifically state that there’s nothing being accused here, and it’s easy to make mistakes with so many blots in a single paper. The comment did not violate PLoS terms.
This is why I love PubPeer so much. So I trotted over there and uploaded my comment. It stayed there in all its DOI-linkable goodness, yielding some interesting responses about how the original comment had disappeared from the PLoS website. Note that you can also opt to notify the authors of a paper when you comment on PubPeer – the authors have not responded so far. I have no idea if PLoS automatically notifies the authors if a comment is left, but if they do then the authors have known about these questions since July 18th.
So what about some metrics? As of today (November 8th), the PubPeer comment has been viewed about 1000 times, and the paper itself about 10,000 times. That means 10% of the people who’ve read this paper at PLoS know about the problems with it. That’s huge! Advertisers would kill for that kind of visibility. PubPeer has most definitely arrived as a format.
I emailed PLoS Biology (the editor who contacted me about the comment) on August 29th (6 weeks gone), asking for a progress report. This time I CC’ed the editor-in-chief. They responded with a boilerplate about how they comply with COPE guidelines.
I emailed PLos Biology again on September 26th (10 weeks). No response.
I emailed PLoS Biology again on October 3rd, asking for a status update. In this email I linked to the PubPeer comment. I also mentioned that Derek Lowe at the amazing “In the Pipeline” blog had written about the paper and its apparent problems. His site gets around 15-20 thousand hits per day. No response from PLoS.
I emailed PLoS Biology again on October 16th with the following…
Nearly 13 weeks (over 3 months) and counting.
I wonder how much longer we will be expected to wait for a resolution to this quite simple case? The courtesy of a response is requested.
PSB
No response. In addition to taking a ridiculously long time to deal with this problem (which thousands of people know about), now they’re just being plain rude and refusing to answer my emails.
So what else can be done? Well thankfully this October saw the launch of PubMed Commons, a shiny new commenting system that’s part of NCBI’s PubMed. Aha! maybe this will work where all else has failed? I happily trotted along and uploaded my comments there. Unfortunately comments are only visible to those logged in via @myNCBI, so if you go to the PubMed page for the paper and log in from there, you should be able to see the comment. Again, I don’t know the inner workings of PubMed Commons, but I would imagine it has to contain some sort of notification system to the journal or the authors. It’s therefore interesting that neither PLoS Biology or the authors have been in touch regarding my comment (using my real name) on PubMed Commons.
As of today (November 8th), it has been 16 weeks since I first raised problems about this paper, and it’s still out there unscathed in the literature, with no indication of any problem whatsoever on PubMed or PLoS Biology’s websites. If I was the pessimistic type, I could take those numbers further up the page and say that 90% of the people reading this paper have no idea about the data problems in it. That’s sad, not to mention dangerous.
Part 3 – What Does it Take?
So here’s the question – what in the name of fuckingcockbuggeringdogshit does it take to get a problem paper dealt with in this day and age?
- I commented on the journal’s own website
- I left comments on PubPeer
- I tweeted about it
- I left comments on PubMed Commons
- It’s been featured on a 15-20k hits/day blog
- I emailed the journal editors until I was blue in the face
- 10% of the people who’ve seen the paper know about the problems
I’m out of options. If a journal can take this much pressure and just brush it all off, what’s the point of all these post publication peer review systems? It should not take this much effort on the part of a reader, to correct the scientific record! It should not take a scientist having to use his own personal lab’ website as the last resort to communicate his frustrations about a totally failed system.
Does anyone agree with me that 16 weeks (nearly 4 months) is too long to establish the authenticity of a few dodgy looking blots?
My lab does a lot of western blotting, and if I could not find you an original film within 24 hours I would be ashamed. I don’t think anyone who calls themselves a real scientist would have trouble finding original data within a couple of days at most. It is completely beyond my understanding why this is taking so long.
Is PLoS Biology really so short-staffed they can’t devote someone to analyze these data for a few hours? PLoS is a $23m/yr. operation; hire more ethics staff already! Do they not have a policy which says “anyone who can’t provide us with original data on demand will have their work retracted”? Don’t they have the ability to put an expression of concern on the paper while it is being investigated? Are they really so rude they can just stop responding to email? Are they really so naive as to think they can ignore the combined power of email, PubPeer, PubMed Commons, Twitter, the blogosphere and at least 10% of an article’s readership? Are they delusional enough to think this problem will disappear if they put on a poker face?
How did we arrive at this totally screwed up system in which the gatekeeper journals decide what the truth is, and their scientist paying customers have to expend enormous personal effort just to get a dodgy looking piece of data explained properly? There has to be a better way.
Finally, I hope it doesn’t need to be spelled out, but the point here is to make a critique on the sorry state of academic publishing, and not to focus on the origins of the problems in the paper itself. There is of course the possibility that I might be wrong. The authors could be sitting on a treasure trove of original data, and they’re going to produce it all for the editors at PLoS Biology to see and this will explain all the anomalies. Maybe a correction will ensue, or maybe PLoS will deem this unnecessary and just close the case. There’s probably no way I’ll ever get to see the original data (that’s not the way these investigations work), so I may just have to take the journal’s word if they say “move along nothing to see”. That’s OK. If I’m wrong on this I promise to apologize profusely for any trouble I’ve caused the authors. Hey, I might even send them a peace offering (authors – if you’re reading this, let me know your beverage of choice).
The bigger point here, is that regardless the outcome of this particular case, it should not take this much time / effort / energy / frustration. The problem is not dodgy looking data, it’s the entire system we have for dealing with it. So don’t focus on this one paper. Instead, recognize that it’s only one example of hundreds that myself and a few like-minded people deal with on a regular basis. It’s a representative case, meant to illustrate a deeper problem.
Thanks for reading.